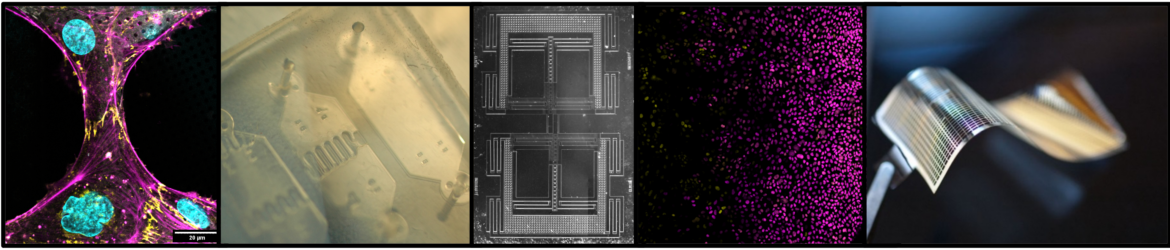
Micropatterning technology for cellular cryo-electron microscopy (cryo-EM) studies
Cryo-electron tomography (cryo-ET) is a 3D cryo-EM technique that is transforming the field of structural biology, and with it medicine, by enabling the visualization of protein structures in the context of intact cells. Single particle cryo-EM has led to a “resolution revolution” in structural biology, earning a Nobel Prize in 2017. However, only recently have advances in instrumentation, sample preparation, and image processing for cryo-ET converged to obtain a previously unsolved protein structure within the context of the cellular environment (Watanabe 2020). As research is ongoing at nearly every stage of the cryo-ET pipeline, standard techniques for cellular sample preparation, data acquisition, and image processing have yet to be established. We will harness cryo-ET to gain insights into the nanometer-scale organization of cell-cell and cell-ECM contacts that underly cell mechanics. The lab will employ micropatterning technologies that Dr. Engel developed in her postdoctoral studies (Engel 2019, Engel 2021) to provide programmed mechanical cues to cells imaged by cryo-ET.
Our first project focuses on endothelial cells. The endothelium lines the vasculature and provides barrier function in an environment subject to large mechanical stresses due to factors such as pulsatile blood flow. Endothelial cells must actively remodel to resist mechanical disruption, as well as to facilitate wound healing and vessel remodeling. While it is known that alterations in the organization of endothelial cell- cell junctions can lead to vascular leakage, inflammation, atherosclerosis, and tumor-associated angiogenesis, the detailed macromolecular organization at these mechanosensitive cell-cell junctions is poorly understood. We will use cryo-ET to elucidate the cytoskeletal and adhesion architectures that allow endothelial cell-cell junctions to be both highly dynamic and mechanically robust.
References:
[1] Watanabe, R. et al. The In situ Structure of Parkinson’s Disease-Linked LRRK2. Biophys. J. 118, 486a (2020) https://doi.org/10.1016/j.cell.2020.08.004. [2] Engel, L. et al. Extracellular matrix micropatterning technology for whole cell cryogenic electron microscopy studies. J. Micromechanics Microengineering (2019) https://doi.org/10.1088/1361-6439/ab419a. [3] Engel, L. et al. Lattice micropatterning for cryo-electron tomography studies of cell-cell contacts. Journal of Structural Biology (2021) https://doi.org/10.1016/j.jsb.2021.107791Microfluidic platforms to enable the patterning of 3D tissue in vitro
A marvel of embryonic development is how uniform populations of stem cells give rise to spatially patterned tissues comprised of multiple cell types. Harnessing the processes by which complex tissues arise during embryonic development would improve our ability to engineer complex, tissue-like constructs in vitro—a longstanding goal of tissue engineering and regenerative medicine. A tenet of developmental biology is that within the embryo, stem cells are exposed to spatial gradients of diffusible extracellular signaling molecules known as morphogens. Graded levels of morphogens induce stem cell differentiation into distinct cell types in a concentration-dependent manner at different positions along the gradient, thus creating spatially patterned tissues. However, many outstanding questions continue to surround the morphogen model due to the experimental challenge of visualizing or quantitatively manipulating morphogen gradients in the context of a live embryo. Specifically, how the information contained in a morphogen gradient is translated into precisely defined tissue architectures remains unclear. To address this and related questions, we will create reductionist microfluidic culture systems that expose human pluripotent stem cells to precisely controlled morphogen gradients that can be predictably modelled using finite element simulations (e.g. COMSOL). We will use time-lapse, live-cell imaging, to characterize the spatial and temporal dynamics of differentiation within the induced signaling gradients. These measurements will reveal how precise patterns of cell types emerge, for example by localized differentiation, or alternatively by noisy differentiation followed by self-segregation of like cell types. In developing a toolset for guiding tissue patterning in an engineering context, we will quantitatively address mechanistic questions in stem cell and developmental biology that are difficult to address in vivo. In addition, these assays will contribute toward the engineering of increasingly realistic tissue-like constructs.
References:
[1] Cui, K.W. and Engel, L., et al. Spatially controlled stem cell differentiation via morphogen gradients: A comparison of static and dynamic microfluidic platforms. Journal of Vacuum Science & Technology A 38, 033205 (2020); https://doi.org/10.1116/1.5142012 [2] Engel, L. and Cui, K.W. et al. A microfluidic platform for anterior-posterior human endoderm patterning via countervailing morphogen gradients in vitro. Under review in iScience.